Malaria parasite detection
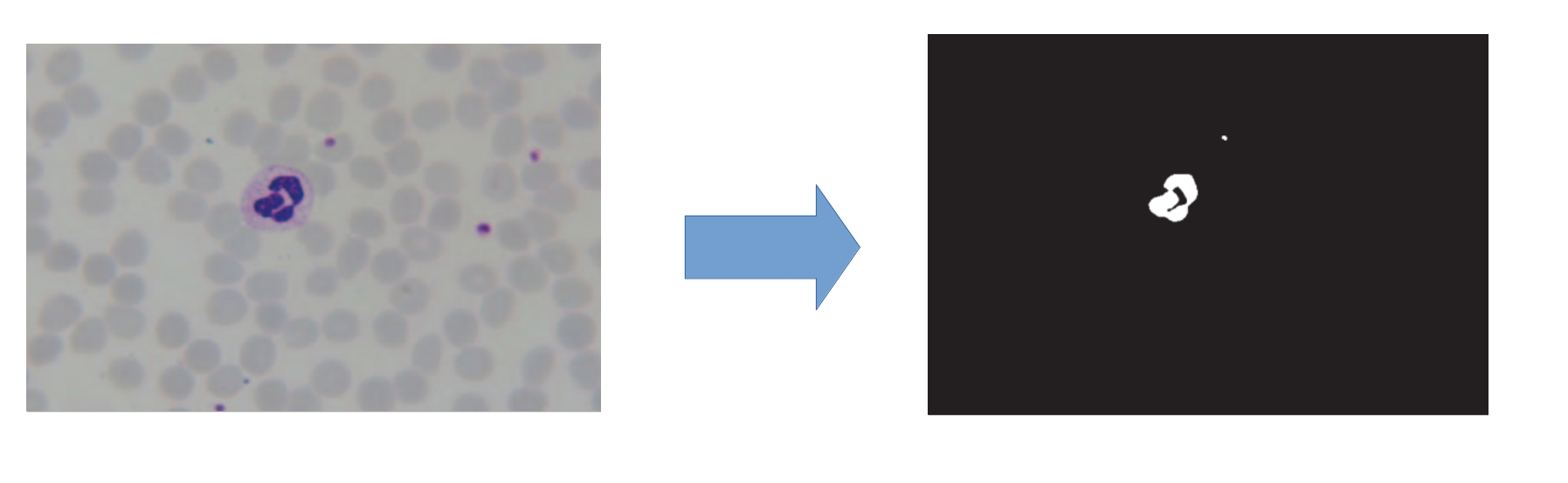 Extracting red blood cell mask, left is the blood cell image, right is red blood cell mask
Extracting red blood cell mask, left is the blood cell image, right is red blood cell mask
This research represents a method to detect malaria parasite in blood samples stained with giemsa. In order to increase the accuracy of detecting, at the first step, the red blood cell mask is extracted. It is due to the fact that most of malaria parasites exist in red blood cells. Then, stained elements of blood such as red blood cells, parasites and white blood cells are extracted. At the next step, red blood cell mask is located on the extracted stained elements to separate the possible parasites. Finally, color histogram, granulometry, gradient and flat texture features are extracted and used as classifier inputs. Here, five classifiers were used: support vector machines (SVM), nearest mean (NM), K nearest neighbors (KNN), 1-NN and Fisher. In this research K nearest neighbors classifier had the best accuracy, which was 91%.
- Different classifires and combination methods applied (Refer to my paper)
Improvement in Classification Accuracy Rate Using Multiple Classifier Fusion
This study was conducted using 400 confirmed images of blood slides infected with malaria parasite. The MATLAB software was used for the implementation of computation procedures. Using five extracted features (flat texture, saturation channel histogram, color histogram, gradient, and granulometry) and six classifiers (k-Nearest Neighbors (k-NN), 1-Nearest Neighbor (1-NN), decision tree (DT), Fisher, linear discriminant analysis (LDA), and quadratic discriminant analysis (QDA)), images were classified into two classes: parasitic and nonparasitic. Then, classifier fusion was done using several algorithms: mean, min, max, stack, median, Adaboost, and bagging.
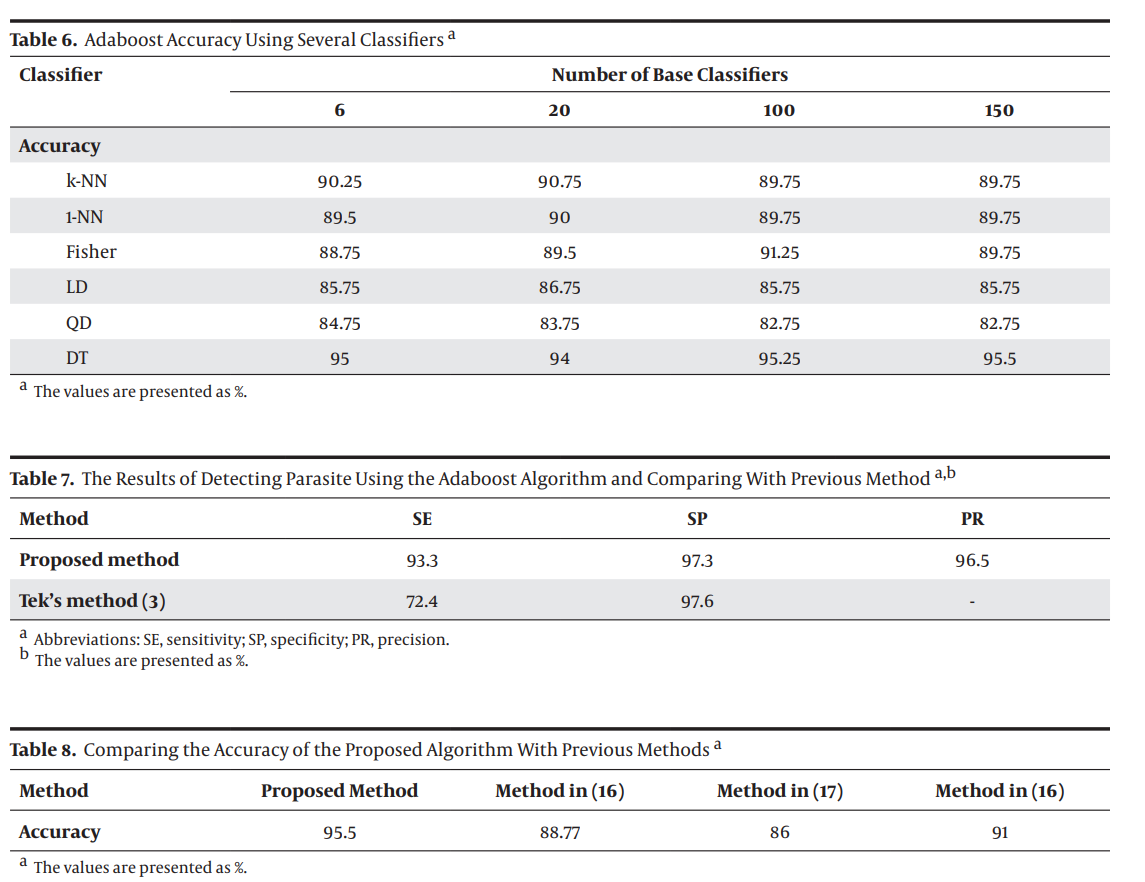 From My paper the results for detecting malaria parasite using Classifier Fusion
From My paper the results for detecting malaria parasite using Classifier Fusion
